 Global Information
Global InformationDNA methylation information

DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.
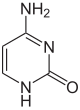
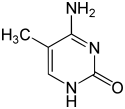
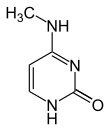
As of 2016, two nucleobases have been found on which natural, enzymatic DNA methylation takes place: adenine and cytosine. The modified bases are N6-methyladenine,[1] 5-methylcytosine[2] and N4-methylcytosine.[3]
| Unmodified base | 
|
|
||||||
| Adenine, A | Cytosine, C | |||||||
| Modified forms | 
|
|
|
|||||
| N6-Methyladenine, 6mA | 5-Methylcytosine, 5mC | N4-Methylcytosine, 4mC | ||||||
Cytosine methylation is widespread in both eukaryotes and prokaryotes, even though the rate of cytosine DNA methylation can differ greatly between species: 14% of cytosines are methylated in Arabidopsis thaliana, 4% to 8% in Physarum,[4] 7.6% in Mus musculus, 2.3% in Escherichia coli, 0.03% in Drosophila; methylation is essentially undetectable in Dictyostelium;[5][6] and virtually absent (0.0002 to 0.0003%) from Caenorhabditis[7] or fungi such as Saccharomyces cerevisiae and S. pombe (but not N. crassa).[8][9]: 3699 Adenine methylation has been observed in bacterial, plant, and recently in mammalian DNA,[10][11] but has received considerably less attention.
Methylation of cytosine to form 5-methylcytosine occurs at the same 5 position on the pyrimidine ring where the DNA base thymine's methyl group is located; the same position distinguishes thymine from the analogous RNA base uracil, which has no methyl group. Spontaneous deamination of 5-methylcytosine converts it to thymine. This results in a T:G mismatch. Repair mechanisms then correct it back to the original C:G pair; alternatively, they may substitute A for G, turning the original C:G pair into a T:A pair, effectively changing a base and introducing a mutation. This misincorporated base will not be corrected during DNA replication as thymine is a DNA base. If the mismatch is not repaired and the cell enters the cell cycle the strand carrying the T will be complemented by an A in one of the daughter cells, such that the mutation becomes permanent. The near-universal use of thymine exclusively in DNA and uracil exclusively in RNA may have evolved as an error-control mechanism, to facilitate the removal of uracils generated by the spontaneous deamination of cytosine.[12] DNA methylation as well as many of its contemporary DNA methyltransferases have been thought to evolve from early world primitive RNA methylation activity and is supported by several lines of evidence.[13]
In plants and other organisms, DNA methylation is found in three different sequence contexts: CG (or CpG), CHG or CHH (where H correspond to A, T or C). In mammals however, DNA methylation is almost exclusively found in CpG dinucleotides, with the cytosines on both strands being usually methylated. Non-CpG methylation can however be observed in embryonic stem cells,[14][15][16] and has also been indicated in neural development.[17] Furthermore, non-CpG methylation has also been observed in hematopoietic progenitor cells, and it occurred mainly in a CpApC sequence context.[18]
- ^ D. B. Dunn, J. D. Smith: "The occurrence of 6-methylaminopurine in deoxyribonucleic acids". In: Biochem J. 68(4), Apr 1958, S. 627–636. PMID 13522672. PMC 1200409.
- ^ B. F. Vanyushin, S. G. Tkacheva, A. N. Belozersky: "Rare bases in animal DNA". In: Nature. 225, 1970, S. 948–949. PMID 4391887.
- ^ Melanie Ehrlich, Miguel A. Gama-Sosa, Laura H. Carreira, Lars G. Ljungdahl, Kenneth C. Kuo, Charles W. Gehrke: "DNA methylation in thermophilic bacteria: N6-methylcytosine, 5-methylcytosine, and N6-methyladenine." In: Nucleic Acids Research. 13, 1985, S. 1399. PMID 4000939. PMC 341080.
- ^ Evans HH, Evans TE (December 1970). "Methylation of the deoxyribonucleic acid of Physarum polycephalum at various periods during the mitotic cycle". The Journal of Biological Chemistry. 245 (23): 6436–6441. doi:10.1016/S0021-9258(18)62627-4. PMID 5530731.

- ^ Smith SS, Rather DI (July 1991). "Lack of 5-methylcytosine in Dictyostelium discoideum DNA". The Biochemical Journal. 277 (1): 273–275. doi:10.1042/bj2770273. PMC 1151219. PMID 1713034.
- ^ Drewell RA, Cormier TC, Steenwyk JL, St Denis J, Tabima JF, Dresch JM, Larochelle DA (April 2023). "The Dictyostelium discoideum genome lacks significant DNA methylation and uncovers palindromic sequences as a source of false positives in bisulfite sequencing". NAR Genomics Bioinformatics. 5 (2): lqad035. doi:10.1093/nargab/lqad035. PMC 10111430. PMID 37081864.
- ^ Hu CW, Chen JL, Hsu YW, Yen CC, Chao MR (January 2015). "Trace analysis of methylated and hydroxymethylated cytosines in DNA by isotope-dilution LC-MS/MS: first evidence of DNA methylation in Caenorhabditis elegans". The Biochemical Journal. 465 (1): 39–47. doi:10.1042/bj20140844. PMID 25299492.
- ^ Bird A (December 2001). "Molecular biology. Methylation talk between histones and DNA". Science's Compass. Science. 294 (5549): 2113–2115. doi:10.1126/science.1066726. hdl:1842/464. PMID 11739943. S2CID 82947750.
As a result of this process, known as repeat-induced point mutation (RIP), the wild-type Neurospora genome contains a small fraction of methylated DNA, the majority of the DNA remaining nonmethylated.

- ^ Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M (April 2014). "Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species". Analytical Chemistry. 86 (8): 3697–3702. doi:10.1021/ac500447w. PMC 4006885. PMID 24640988.
- ^ Ratel D, Ravanat JL, Berger F, Wion D (March 2006). "N6-methyladenine: the other methylated base of DNA". BioEssays. 28 (3): 309–315. doi:10.1002/bies.20342. PMC 2754416. PMID 16479578.
- ^ Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. (April 2016). "DNA methylation on N(6)-adenine in mammalian embryonic stem cells". Nature. 532 (7599): 329–333. Bibcode:2016Natur.532..329W. doi:10.1038/nature17640. PMC 4977844. PMID 27027282.
- ^ Angéla Békési and Beáta G Vértessy "Uracil in DNA: error or signal?"
- ^ Rana AK, Ankri S (2016). "Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases". Frontiers in Genetics. 7: 99. doi:10.3389/fgene.2016.00099. PMC 4893491. PMID 27375676.
- ^ Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E (May 2002). "De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation". Gene. 289 (1–2): 41–48. doi:10.1016/S0378-1119(02)00469-9. PMID 12036582.
- ^ Haines TR, Rodenhiser DI, Ainsworth PJ (December 2001). "Allele-specific non-CpG methylation of the Nf1 gene during early mouse development". Developmental Biology. 240 (2): 585–598. doi:10.1006/dbio.2001.0504. PMID 11784085.
- ^ Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. (November 2009). "Human DNA methylomes at base resolution show widespread epigenomic differences". Nature. 462 (7271): 315–322. Bibcode:2009Natur.462..315L. doi:10.1038/nature08514. PMC 2857523. PMID 19829295.
- ^ Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. (August 2013). "Global epigenomic reconfiguration during mammalian brain development". Science. 341 (6146): 1237905. doi:10.1126/science.1237905. PMC 3785061. PMID 23828890.
- ^ Kulis M, Merkel A, Heath S, Queirós AC, Schuyler RP, Castellano G, et al. (July 2015). "Whole-genome fingerprint of the DNA methylome during human B cell differentiation". Nature Genetics. 47 (7): 746–756. doi:10.1038/ng.3291. PMC 5444519. PMID 26053498.