 Global Information
Global InformationReverse gyrase information
| Reverse gyrase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
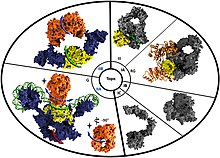 Molecular depictions of the different type I and type II topoisomerase classes. Reverse gyrase is indicated by the 'RG' tag under the type IA topoisomerase section of the diagram. | |||||||||
| Identifiers | |||||||||
| EC no. | 5.6.2.2 | ||||||||
| CAS no. | 143180-75-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Reverse gyrase is a type I topoisomerase that introduces positive supercoils into DNA,[1] contrary to the typical negative supercoils introduced by the type II topoisomerase DNA gyrase. These positive supercoils can be introduced to DNA that is either negatively supercoiled or fully relaxed.[2] Where DNA gyrase forms a tetramer and is capable of cleaving a double-stranded region of DNA, reverse gyrase can only cleave single stranded DNA.[3][4] More specifically, reverse gyrase is a member of the type IA topoisomerase class; along with the ability to relax negatively or positively supercoiled DNA[5] (which does not require ATP[6][7]), type IA enzymes also tend to have RNA-topoisomerase activities. These RNA topoisomerases help keep longer RNA strands from becoming tangled in what are referred to as "pseudoknots." Due to their ability to interact with RNA, it is thought that this is one of the most ancient class of enzymes found to date.[4]
Reverse gyrase is an ATP-dependent topoisomerase[8] in terms of its positive supercoiling activity, however, reverse gyrase can also relax DNA strands without introducing positive supercoils through interaction with ADP.[9] The structure of the enzyme includes both a helicase domain, which is responsible for separating nucleic acids, and a topoisomerase domain, which is responsible for the actual introduction of coils into DNA. However, mechanistic studies have shown that these two domains tend to exhibit weak activities separately and can only perform efficient DNA positive supercoiling activity when working in tandem.[10][11] Other studies have also shown that reverse gyrase enzymes tend to favorably attack regions of single-stranded DNA versus double-stranded DNA, which suggests that this enzyme's critical biological function is to ensure the constant renaturation of melted DNA strands, especially in organisms that grow at high temperatures.[8]
This enzyme has been extensively characterized across several Archaea, with Sulfolobus acidocaldarius reverse gyrase being one of the first to be characterized.[12] Additionally, it has been found that all thermophilic bacteria and archaea contain at least one reverse gyrase enzyme. Some organisms, such as members of the Crenarchaeota phylum, even have two reverse gyrase enzymes: TopR1, which tends to be active in increased temperatures, and TopR2, which shows activity in both low and high temperatures.[4] Other exceptional organisms include Nanoarchaeum equitans, whose reverse gyrase enzyme tends to naturally exist as two separate peptides versus the typical monomeric polypeptide with a topoisomerase IA domain and a helicase domain.[4][11]
- ^ Kikuchi A, Asai K (21 June 1984). "Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA". Nature. 309 (5970): 677–681. Bibcode:1984Natur.309..677K. doi:10.1038/309677a0. PMID 6328327. S2CID 4242694.
- ^ Cite error: The named reference
:6was invoked but never defined (see the help page). - ^ Jaxel C, Nadal M, Mirambeau G, Forterre P, Takahashi M, Duguet M (October 1989). "Reverse gyrase binding to DNA alters the double helix structure and produces single-strand cleavage in the absence of ATP". The EMBO Journal. 8 (10): 3135–3139. doi:10.1002/j.1460-2075.1989.tb08466.x. PMC 401394. PMID 2555155.
- ^ a b c d Garnier F, Couturier M, Débat H, Nadal M (25 May 2021). "Archaea: A Gold Mine for Topoisomerase Diversity". Frontiers in Microbiology. 12: 661411. doi:10.3389/fmicb.2021.661411. PMC 8185306. PMID 34113328.
- ^ Cite error: The named reference
:4was invoked but never defined (see the help page). - ^ "EC 5.6.2.1". IUBMB Enzyme Nomenclature. International Union of Biochemistry and Molecular Biology.
- ^ Gellert M (1981). "DNA topoisomerases". Annual Review of Biochemistry. 50: 879–910. doi:10.1146/annurev.bi.50.070181.004311. PMID 6267993.
- ^ a b Shibata T, Nakasu S, Yasui K, Kikuchi A (August 1987). "Intrinsic DNA-dependent ATPase activity of reverse gyrase". The Journal of Biological Chemistry. 262 (22): 10419–10421. doi:10.1016/S0021-9258(18)60974-3. PMID 3038879.
- ^ Cite error: The named reference
:5was invoked but never defined (see the help page). - ^ Déclais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M (June 2000). "Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling". The Journal of Biological Chemistry. 275 (26): 19498–19504. doi:10.1074/jbc.M910091199. PMID 10748189.
- ^ a b Capp C, Qian Y, Sage H, Huber H, Hsieh TS (December 2010). "Separate and combined biochemical activities of the subunits of a naturally split reverse gyrase". The Journal of Biological Chemistry. 285 (51): 39637–39645. doi:10.1074/jbc.M110.173989. PMC 3000944. PMID 20929866.
- ^ Lulchev P, Klostermeier D (July 2014). "Reverse gyrase--recent advances and current mechanistic understanding of positive DNA supercoiling". Nucleic Acids Research. 42 (13): 8200–8213. doi:10.1093/nar/gku589. PMC 4117796. PMID 25013168.