 Global Information
Global InformationCirculating tumor DNA information
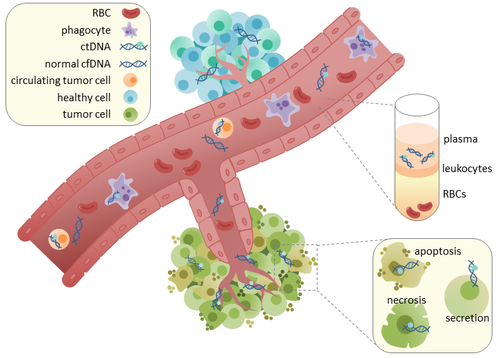
Circulating tumor DNA (ctDNA) is tumor-derived fragmented DNA in the bloodstream that is not associated with cells. ctDNA should not be confused with cell-free DNA (cfDNA), a broader term which describes DNA that is freely circulating in the bloodstream, but is not necessarily of tumor origin. Because ctDNA may reflect the entire tumor genome, it has gained traction for its potential clinical utility; "liquid biopsies" in the form of blood draws may be taken at various time points to monitor tumor progression throughout the treatment regimen.[1][2]
Recent studies have laid the foundation for inferring gene expression from cfDNA (and ctDNA), with EPIC-seq emerging as a notable advancement.[3] This method has substantially raised the bar for the noninvasive inference of expression levels of individual genes, thereby augmenting the assay's applicability in disease characterization, histological classification, and monitoring treatment efficacy.[3][4][5]
ctDNA originates directly from the tumor or from circulating tumor cells (CTCs),[6] which describes viable, intact tumor cells that shed from primary tumors and enter the bloodstream or lymphatic system. The precise mechanism of ctDNA release is unclear. The biological processes postulated to be involved in ctDNA release include apoptosis and necrosis from dying cells, or active release from viable tumor cells.[7][8][9][10][11] Studies in both human (healthy and cancer patients)[12] and xenografted mice[13] show that the size of fragmented cfDNA is predominantly 166bp long, which corresponds to the length of DNA wrapped around a nucleosome plus a linker. Fragmentation of this length might be indicative of apoptotic DNA fragmentation, suggesting that apoptosis may be the primary method of ctDNA release. The fragmentation of cfDNA is altered in the plasma of cancer patients.[14][15] In healthy tissue, infiltrating phagocytes are responsible for clearance of apoptotic or necrotic cellular debris, which includes cfDNA.[16] ctDNA in healthy patients is only present at low levels but higher levels of ctDNA in cancer patients can be detected with increasing tumor sizes.[17] This possibly occurs due to inefficient immune cell infiltration to tumor sites, which reduces effective clearance of ctDNA from the bloodstream.[16] Comparison of mutations in ctDNA and DNA extracted from primary tumors of the same patients revealed the presence of identical cancer-relevant genetic changes.[18][19] This led to the possibility of using ctDNA for earlier cancer detection and treatment follow up.[20]
- ^ Wan J, Massie C, Garcia-Corbacho J, Mouliere F, Brenton J, Caldas C, Pacey S, Baird R, Rosenfeld N (April 2017). "Liquid biopsies come of age: towards implementation of circulating tumour DNA". Nature Reviews Cancer. 17 (4): 223–238. doi:10.1038/nrc.2017.7. PMID 28233803. S2CID 4561229.
- ^ Nonaka, T; Wong, DTW (13 June 2022). "Saliva Diagnostics". Annual Review of Analytical Chemistry. 15 (1): 107–121. Bibcode:2022ARAC...15..107N. doi:10.1146/annurev-anchem-061020-123959. PMC 9348814. PMID 35696523.
- ^ a b Esfahani, Mohammad Shahrokh; Hamilton, Emily G.; Mehrmohamadi, Mahya; et al. (April 2022). "Inferring gene expression from cell-free DNA fragmentation profiles". Nature Biotechnology. 40 (4): 585–597. doi:10.1038/s41587-022-01222-4. PMC 9337986. PMID 35361996.
- ^ Mutter, Jurik A; Shahrokh Esfahani, Mohammad; Schroers-Martin, Joseph; et al. (28 November 2023). "Inferred Gene Expression By Cell-Free DNA Profiling Allows Noninvasive Lymphoma Classification". Blood. 142 (Supplement 1): 245–245. doi:10.1182/blood-2023-186853.
- ^ Alig, Stefan K.; Shahrokh Esfahani, Mohammad; Garofalo, Andrea; et al. (25 January 2024). "Distinct Hodgkin lymphoma subtypes defined by noninvasive genomic profiling". Nature. 625 (7996): 778–787. doi:10.1038/s41586-023-06903-x. PMID 38081297.
- ^ Akca H, Demiray A, Yaren A, Bir F, Koseler A, Iwakawa R, Bagci G, Yokota J (March 2013). "Utility of serum DNA and pyrosequencing for the detection of EGFR mutations in non-small cell lung cancer". Cancer Genetics. 206 (3): 73–80. doi:10.1016/j.cancergen.2013.01.005. PMID 23491080.
- ^ Schwarzenbach H, Hoon DS, Pantel K (June 2011). "Cell-free nucleic acids as biomarkers in cancer patients". Nature Reviews. Cancer. 11 (6): 426–37. doi:10.1038/nrc3066. PMID 21562580. S2CID 6061607.
- ^ Stroun M, Anker P (July 1972). "Nucleic acids spontaneously released by living frog auricles". The Biochemical Journal. 128 (3): 100P–101P. doi:10.1042/bj1280100pb. PMC 1173871. PMID 4634816.
- ^ Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P (November 2001). "About the possible origin and mechanism of circulating DNA apoptosis and active DNA release". Clinica Chimica Acta; International Journal of Clinical Chemistry. 313 (1–2): 139–42. doi:10.1016/S0009-8981(01)00665-9. PMID 11694251.
- ^ Anker P, Stroun M, Maurice PA (September 1975). "Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system". Cancer Research. 35 (9): 2375–82. PMID 1149042.
- ^ Rogers JC, Boldt D, Kornfeld S, Skinner A, Valeri CR (July 1972). "Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen". Proceedings of the National Academy of Sciences of the United States of America. 69 (7): 1685–9. Bibcode:1972PNAS...69.1685R. doi:10.1073/pnas.69.7.1685. PMC 426778. PMID 4505646.
- ^ Heitzer E, Auer M, Hoffmann EM, Pichler M, Gasch C, Ulz P, Lax S, Waldispuehl-Geigl J, Mauermann O, Mohan S, Pristauz G, Lackner C, Höfler G, Eisner F, Petru E, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl JB, Speicher MR (July 2013). "Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer". International Journal of Cancer. 133 (2): 346–56. doi:10.1002/ijc.28030. PMC 3708119. PMID 23319339.
- ^ Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Del Rio M, Molina F (October 2010). "Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts". Nucleic Acids Research. 38 (18): 6159–75. doi:10.1093/nar/gkq421. PMC 2952865. PMID 20494973.
- ^ Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, Gongora C, Thierry AR (2011). "High fragmentation characterizes tumour-derived circulating DNA". PLOS ONE. 6 (9): e23418. Bibcode:2011PLoSO...623418M. doi:10.1371/journal.pone.0023418. PMC 3167805. PMID 21909401.
- ^ Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. (November 2018). "Enhanced detection of circulating tumor DNA by fragment size analysis". Sci Transl Med. 10 (466). doi:10.1126/scitranslmed.aat4921. PMC 6483061. PMID 30404863.
- ^ a b Pisetsky DS, Fairhurst AM (June 2007). "The origin of extracellular DNA during the clearance of dead and dying cells". Autoimmunity. 40 (4): 281–4. doi:10.1080/08916930701358826. PMID 17516210. S2CID 11499768.
- ^ Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, Alizadeh AA, Diehn M, Reiter JG (December 2020). "A mathematical model of ctDNA shedding predicts tumor detection size". Science Advances. 6 (50): eabc4308. Bibcode:2020SciA....6.4308A. doi:10.1126/sciadv.abc4308. PMC 7732186. PMID 33310847. S2CID 228096858.
- ^ Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M (April 1994). "Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia". British Journal of Haematology. 86 (4): 774–779. doi:10.1111/j.1365-2141.1994.tb04828.x. PMID 7918071. S2CID 26365875.
- ^ Vasioukhin V, Stroun M, Maurice P, Lyautey J, Lederrey C, Anker P (May 1994). K-ras point mutations in the blood plasma DNA of patients with colorectal tumors in Challenges of Modern Medicine. Vol. 5. pp. 141–150.
- ^ Yong E (July 2014). "Cancer biomarkers: Written in blood". Nature. 511 (7511): 524–526. Bibcode:2014Natur.511..524Y. doi:10.1038/511524a. PMID 25079538. S2CID 4445938.